Research
Detection of Epigenetic Traits from Next Generation Sequencing Data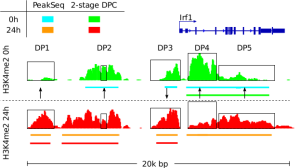 We develop methods for detection of epigenetic features of cell, such as cell active binding sites from open chromatin essays and chromatin changes during biological processes. These methods include the integrative analysis of NGS data as DNase-, ChIP-, and RNA-Seq and single cell variants of these protocols. In this context, we develop a toolbox to perform several basic computation tasks required for the analysis of regulatory genomics data. The toolbox also includes motif matching from DNA sequences, signal processing of next generation sequencing, manipulation and graphical analysis of genomic intervals and profiles and is available at www.regulatory-genomics.org . |
Relating Expression/Genotypes to Diseases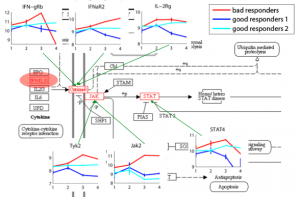 Gene expression and DNA methylation profiling allows clinical diagnosis to be made on a molecular level, thereby substantially increasing diagnosis accuracy and facilitating choice of treatment based on the patients’ genetic traits. Moreover, identifying disease-related genes and monitoring their activity levels provide insights into disease mechanisms. We aim to improve methods for disease diagnosis and marker selection by integrating additional sources of data and the design of methods capable of handling high dimensional spaces. |
Regulation via long non-coding RNA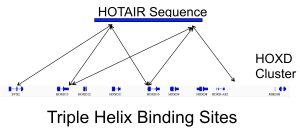 RNA sequencing indicates that a majority of transcripts expressed in one cell type is non-coding. Particularly interesting are the long non-coding RNAs (lncRNA), which can combine a myriad of functional domain characteristics of RNA molecules to act as modular scaffolds promoting the interaction of several proteins, RNA and DNA molecules. We are particularly interested in the triple helices—the interaction of RNA with DNA double helices. We currently develop algorithms for detection of triple helices and investigate regulatory roles of lncRNAs differentially expressed during cell differentiation and diseases. |
Transcriptional regulation and epigenetics during cell differentiation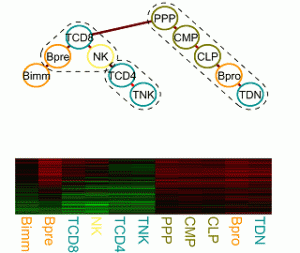
The regulatory processes that govern cell proliferation and differentiation are central to developmental biology. Particularly well studied in this respect is the hematopoietic system. Gene expression, regulation and epigenetic data of cells of various distinguishable developmental stages foster the elucidation of the underlying molecular processes, which change gradually over time and lock cells in certain lineages or induce cell potency. We are interested in developing a statistical framework for tasks as finding modules of functionally related genes and characterizing cell potency and commitment from expression and epigenetic signatures. |
Dynamics of Gene Expression and Function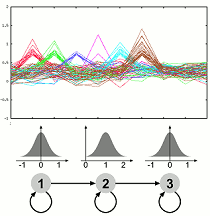 The molecular processes of life are dynamic over time. Experiments measuring the expression levels of a multitude of genes over time are one way of gaining insight into the dynamic processes. We have developed an approach which allows to use prior knowledge, is flexible and very robust to noise. The method is implemented in the software GQL which allows control of the analysis process by use of graphical user interfaces. We are currently extending our framework to allow integration of further data related to transcription or protein interactions and investigating methodologies for validating clustering of genes with functional annotation.
|
