Research
Understanding Cellular Dynamics in Health and Disease
We develop computational methods to uncover how cells change their identity, interact within tissues, and contribute to human disease. We integrate single-cell, spatial, and epigenomic data with advanced statistical and machine-learning approaches to reconstruct dynamic biological processes. Our goal is to reveal the molecular programs and cellular ecosystems that drive pathology — and to provide tools that empower the broader scientific community. Our work spans four interconnected research themes:
Computational Methods for Analysis of Single Cell and Spatial Disease Atlas
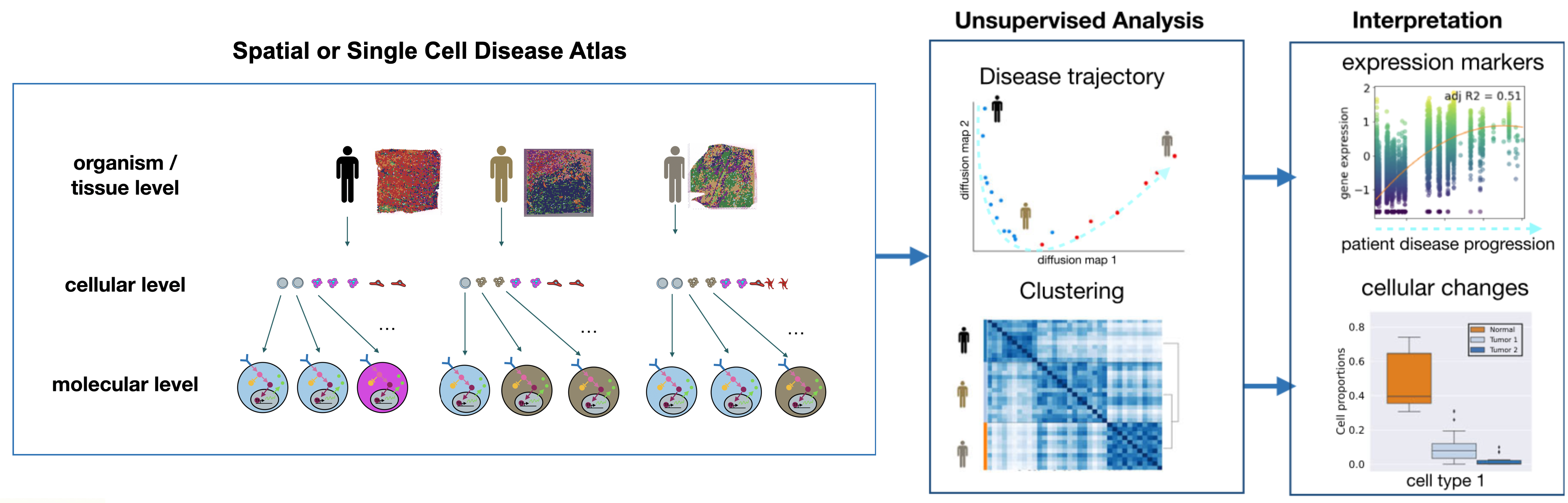
We build statistical and machine-learning tools to analyze and integrate multi-scale omics data, from single-cell transcriptomics and epigenomics to spatial tissue maps and clinical phenotypes. A central focus is our PILOT framework, which uses optimal transport theory to model continuous disease progression across individuals. This approach allows us to reconstruct disease trajectories, identify pivotal molecular events, and map how tissue composition evolves over time (https://diseasecellatlas.org/). This has been used to uncover molecular mechanism driving fibrosis in kidney and heart diseases.
Dissecting cell differentiation and gene regulatory process
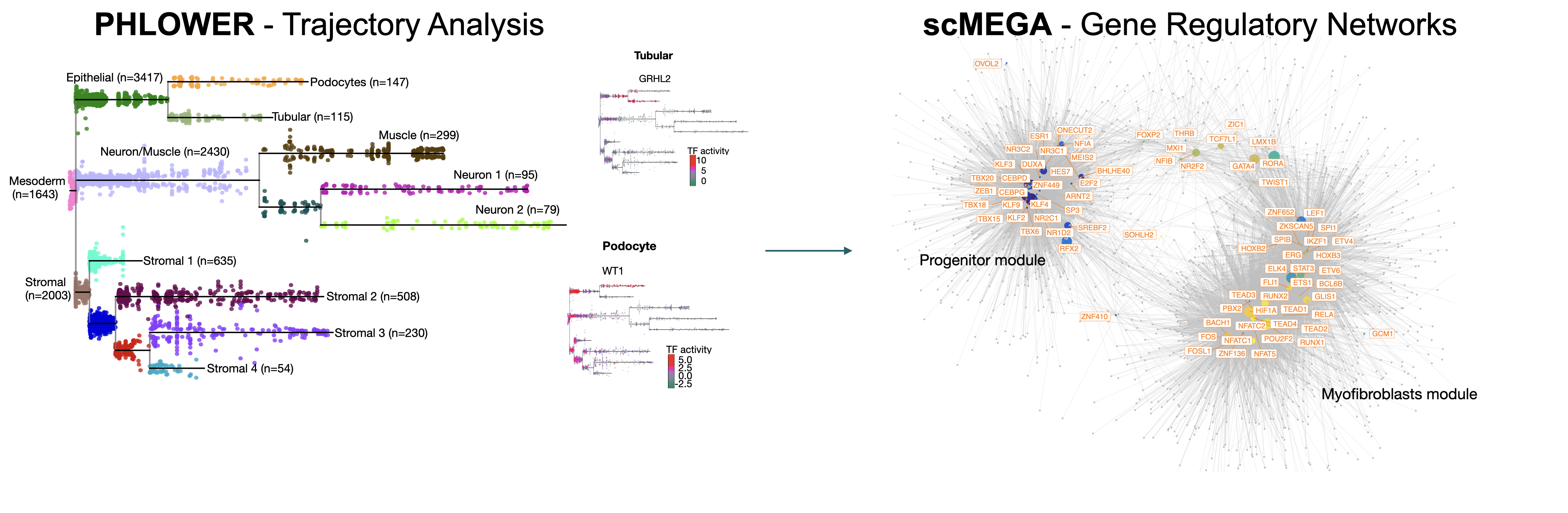
We develop computational methods that explain how cells acquire distinct identities through coordinated changes in gene expression, chromatin accessibility, and transcription factor activity. Our algorithms include PHLOWER, a topology-aware trajectory inference method that reconstructs continuous cell-state transitions, and scMEGA, a framework that integrates single-cell transcriptomic and epigenomic measurements to infer enhancer-driven regulatory networks. We also created HINT, a digital footprinting suite that identifies transcription factor binding sites and regulatory activity from chromatin accessibility data (ATAC-seq). These tools enable mechanistic studies of differentiation and cell-state transitions across multiple biological contexts. For example, we investigate how kidney organoids can be guided toward more mature states, how regulatory programs driving fibroblasts in post-infarction cardiac repair, and how HNF6 variants affects pancreas cell differentiation causing juvenile diabetes.
Uncovering spatial biology and cell-cell communication in diseases
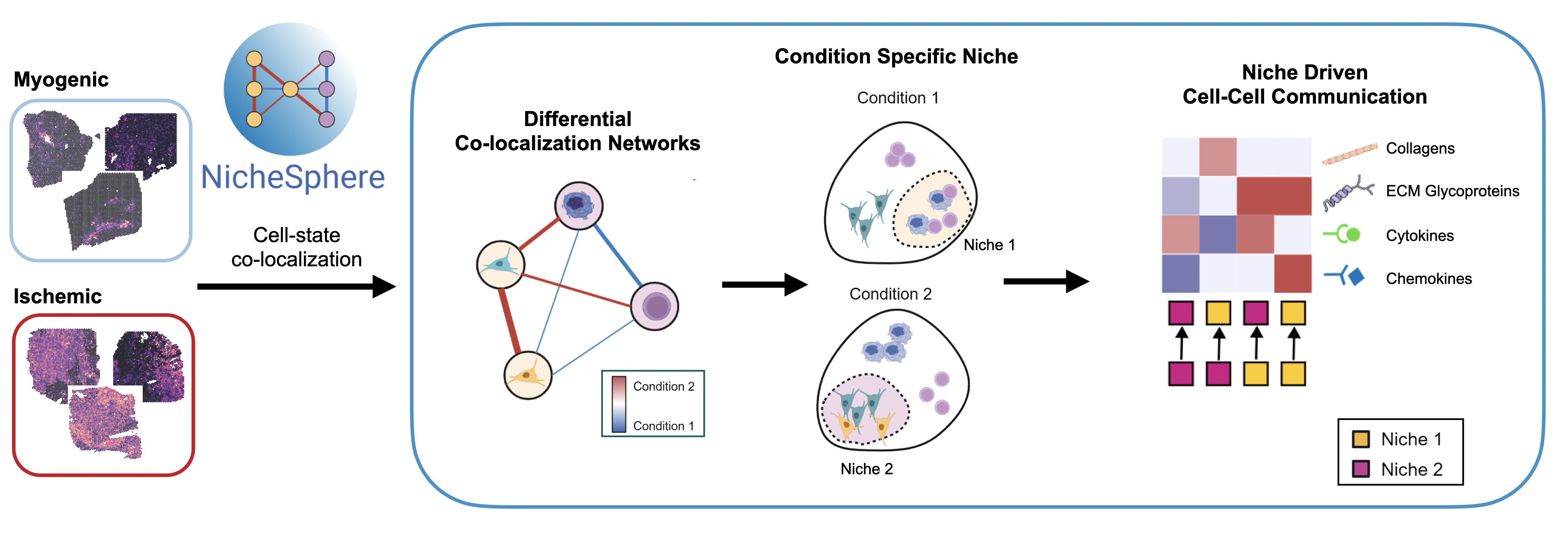
Understanding how cells behave within their tissue environment is essential for deciphering complex diseases. We develop computational methods that combine single cell and spatial transcriptomics with models of intercellular communication. CrossTalkeR allows us to identify and contrast ligand–receptor interactions across conditions such as healthy and diseased tissues, highlighting how communication networks are rewired in pathology. NicheSphere expands these capabilities by modeling co-localization data from spatial transcriptomics data as graphs, allowing us to define and analyze tissue niches — localized communities of interacting cells whose coordinated behavior drives biological and pathological processes.
Using these tools, we map how cell–cell communication shifts during disease progression and identify intercellular signaling mechanisms that may represent new therapeutic targets. For example, cell–cell communication analysis in bone-marrow myelofibrosis revealed an immune-driven rewiring of fibroblast signaling via alarmins. A drug targeting these alarmins is now being repurposed and tested in a clinical trial, representing a clear illustration of how single-cell computational insights can translate to bench and bedside advances
Detection of Epigenetic traits from Sequencing Data
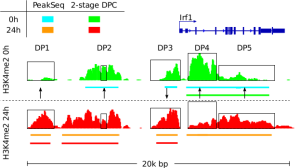
Epigenetic architecture determines how cells interpret their genomes and respond to regulatory signals. We study how chromatin accessibility, transcription factor binding, and enhancer activity shape cellular behavior across developmental and disease contexts. Our methods detect regulatory DNA features from ATAC-seq, ChIP-seq, and single-cell epigenomic data; track regulatory activity through transcription factor footprints; and integrate epigenomic signatures with gene expression and chromatin state transitions.
These efforts are supported by the Regulatory Genomics Toolbox, a software suite that offers motif matching, transcription factor annotation, sequencing signal processing, genomic interval and profile manipulation, and high-quality visualization tools (www.regulatory-genomics.org).
