Detecting differential peaks in ChIP-seq signals with ODIN
Manuel Allhoff, Heike Chauvistré, Kristin Seré, Qiong Lin, Martin Zenke and Ivan G. Costa
Method
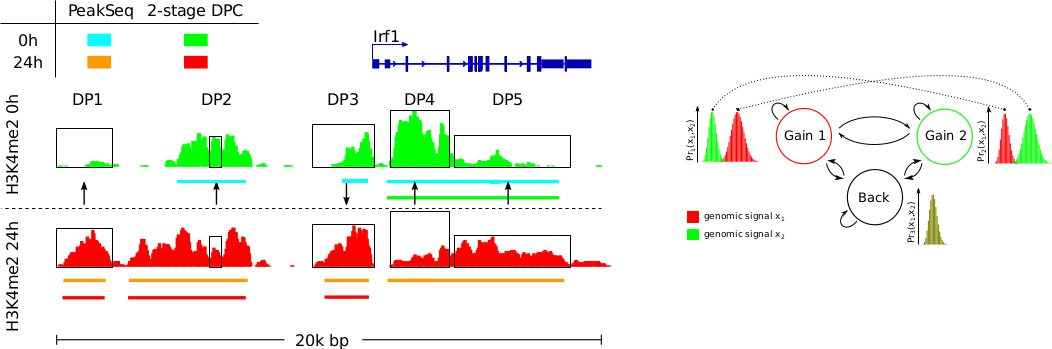
Detection of changes in DNA-protein interactions under distinct cellular conditions from ChIP-seq data is a crucial step in unravelling the regulatory networks behind biological processes. The simplest variation of this problem, which we call differential peak calling, is defined as finding a genomic region where the interaction of a particular protein with DNA is changing between two cellular conditions.
The great majority of peak calling methods can only analyse one ChIP-seq signal at a time and are unable to perform differential peak calling. Recently, a few approaches, based on the combination of these peak callers with statistical tools, have been proposed to detect differential digital gene expression. However, these methods fail to detect detailed changes of protein-DNA interactions.
We propose ODIN; a HMM-based approach to detect and analyse differential peaks in pairs of ChIP-seq data. ODIN is the first differential peak caller that performs genomic signal processing, peak calling and p-value calculation in an integrated framework. We also propose an evaluation methodology to compared ODIN with competing methods. It is based on the association of differential peaks with expression changes in the same cellular conditions. Our empirical study based on 15 ChIP-seq experiments from both transcription factors and histone modifications shows that ODIN outperforms all competing methods.